Designing QPCR oligos might seem complicated but there are some rules and softwares that can make it easy. In our lab, majority of data points we generate are probably measured by QPCR, so I think it is worth to review an algorithm for desiging QPCR assays.
So, today I will describe the way I design QPCR oligos. You can have a basic intro in PCR here.
More than a year ago I switched to the UPL system, the library of probes designed by Exiqon and now marketed by Roche. The concept is quite clear, the LNA modified oligonucleotides bind much stronger to the DNA template than the average oligonucleotides. By this we can decrease the lengths of them by keeping the Tm unchanged. The UPL library consists of 165 individual oligonucleotides that are in general nine basepair long and together cover the entire genome in respect of the coverage needed to design a QPCR oligo set for any gene. You can read more about the LNA nucleotides here and here. A good website where you can calculate the Tm of the LNA oligos here: http://lna-tm.com/. The UPL system is described here.
The system allows the design of an oligo set for any DNA sequence in the UPL Design Centre. You can access the Design Centre directly here.

UPL Assay Design Center
Before we design an assay let us first look for the transcripts of a specific gene. It is very important to use annotated data, since in the annotated genomic data we have informations about possible SNP variations. This might be important, since the oligos and especially the probe should bind to an SNP free free region, because an SNP might disturb the binding of the probe to the template.
To make this data available we should use not the sequence but the transcript ID from the Ensembl.

Ensembl
At Ensembl select your species of interest, e.g. mouse and write the name of your gene of interest in the search box:
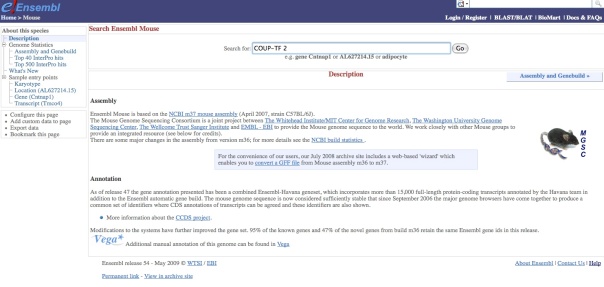
If you write into the box a gene of interest (e.g. COUP-TF2) we will see the results as it is shown here:

Here I have to click on the link of the gene and I will find the following screen:  The most important info we are looking for is in the table on the top of the page:
The most important info we are looking for is in the table on the top of the page:

The two transcripts of the gene are:
ENSMUST00000089565
ENSMUST00000032768.
We will use these ID-s in the UPL Assay Design Centre. First select as organism the “Mouse”, and write into the box the two Ensembl transcript ID-s selected by commas.  If you follow the steps the results will be like this:
If you follow the steps the results will be like this:
 Below this data you can see two links as it follows:
Below this data you can see two links as it follows: To design an assay that would measure all transcripts select: “common assays”.
To design an assay that would measure all transcripts select: “common assays”.
The results will be given in a downloadable pdf report. Save this file and name is by the name of the gene you used as input.
The results in the pdf file look like this:

You can see that the amplicon is 95 bp long, there is no SNP in the binding regions of the primers and probes and the probe is closer to one of the primers. There is an SNP in the gene that was avoided by the program. You can have an SNP even in the amplicon, unless is not in the binding site of the primers or the probe.
Before I order the oligos, I usually test them with e-PCR on the UCSC Genome Browser.

UCSC Genome Browser
Select the PCR view and paste the oligos into the given locations. Select the genome, the assembly and the target as “UCSC Genes”(If you used a genomic sequence for design use the target: “genome assembly”).
 If you have a hit, click on the link provided and the results will be represented in the genome as seen here:
If you have a hit, click on the link provided and the results will be represented in the genome as seen here:

The oligos are intron spanning and in the right location. Order the oligos in an HPLC pufied form in the lowest available scale for the first try. Be aware that according to the experience of several groups, and my own experience too, only 2/3 of the UPL assays work without further optimization. This means, if you want to be sure from the first you better try two, three different assays for the same gene. The UPL Design Centre will generate several primer-probe sets and you can retrive these results too. Since the UPL library is given and one or two of the three ordered assays will work, it this worth trying three from the beginning!
In general the rules for a good QPCR assay:
1. The amplicon should be as short as possible (60-70 bp is ideal, but should be shorter than 100bp).
2. The Tm of the oligos should be around 60C, while the Tm of the probe 10C higher.
3. The distance between the the oligo and the probe should be as small as possible for a better exonuclease activity of the Taq polymerase.
4. The GC content of the two oligos should be as close as possible.
5. The number of GC bases in the last five nucleotides on the 3′ ends of the two primers should be identical (if possible).
6. Select for oligo sets with week internal bonds (less than four H bonds in the same conformation).
7. Avoid primer dimers that could produce artefacts due to the 3′ elongation of one of the primers. The same for internal conformations. Test it on the IDT webpage here. See below:
8. Verify the oligos with e-PCR on the UCSC Genome Browser. The test should give one single hit!
9. If possible use annotated sequences to avoid the SNP effect.
10. If you are looking for genes (cDNA measurement) use exons that are common for all transcripts variants (or use the “batch assay-common assays” in the UPL Assay Design Centre)
Good luck!




 The most important info we are looking for is in the table on the top of the page:
The most important info we are looking for is in the table on the top of the page:
 If you follow the steps the results will be like this:
If you follow the steps the results will be like this: Below this data you can see two links as it follows:
Below this data you can see two links as it follows: To design an assay that would measure all transcripts select: “common assays”.
To design an assay that would measure all transcripts select: “common assays”.

 If you have a hit, click on the link provided and the results will be represented in the genome as seen here:
If you have a hit, click on the link provided and the results will be represented in the genome as seen here:

